Help
Do you have a question regarding or related to the database?
Feel free to
drop us an e-mail (please put
[BiG-FAM HELP] on the subject).
Be sure to first check these
Frequently Asked Questions
to see if your question is already answered there.
Purpose of the database
Example use cases
Exploring rhantipeptides BGC diversity
GCF analysis on a newly sequenced Streptomyces
Frequently Asked Questions (FAQs)
How were the GCFs hosted in BiG-FAM calculated?
How do I compare my own BGC against the GCFs in BiG-FAM?
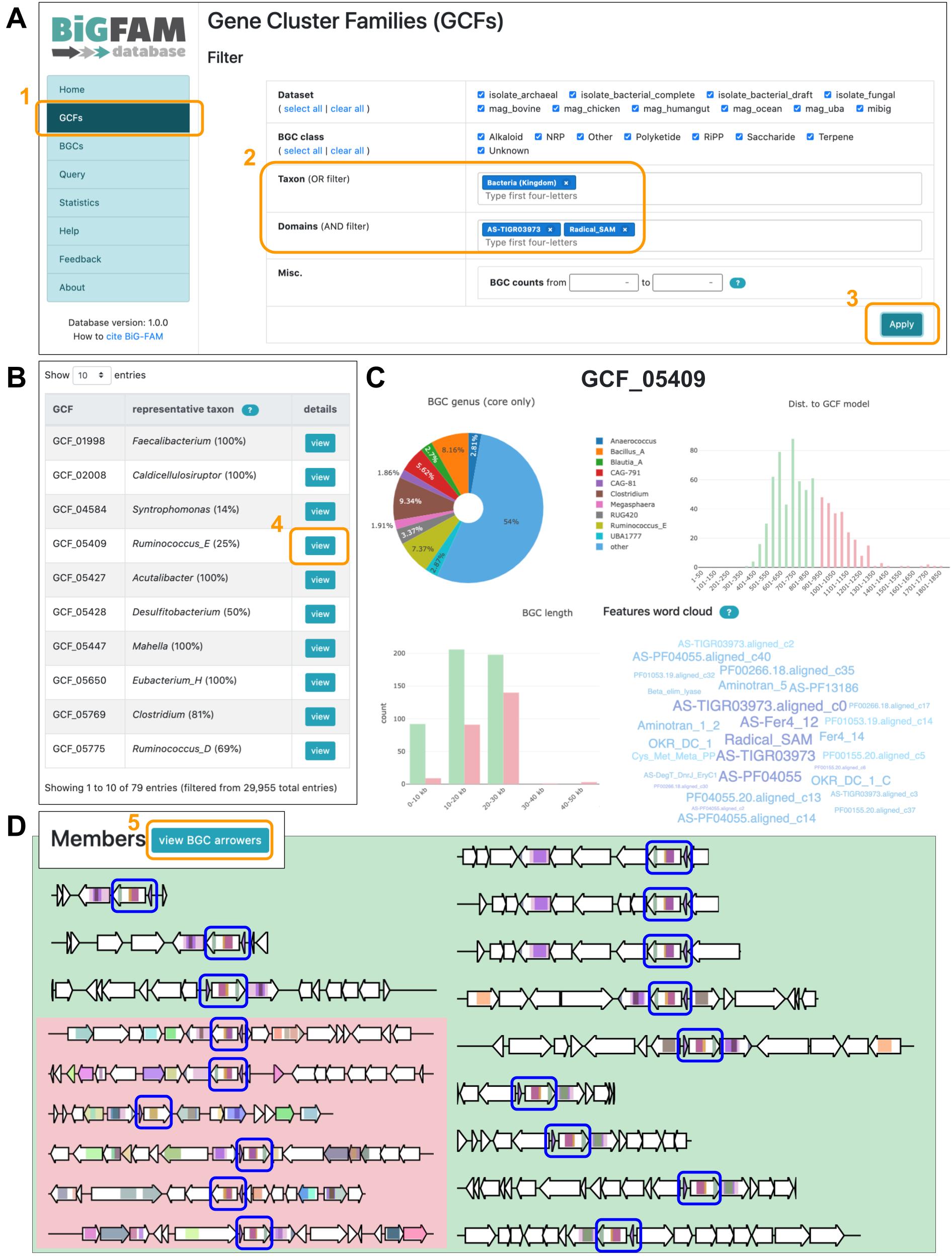
How do I search for BGCs or GCFs with certain characteristics (protein domains, taxonomy, etc.)?
How is BiG-FAM related to antiSMASH, MIBiG, BiG-SCAPE and BiG-SLiCE?
Can I set up a copy of this database on my own (local) servers?
What is the privacy policy of antiSMASH concerning the sequence data used for query mode?
From which studies were the genomes and MAGs used in BiG-FAM sourced?
How do I (bulk) download GCF data from BiG-FAM?
How do I cite BiG-FAM?